
For nucleotide sequences, a similar gap penalty is used, but a much simpler substitution matrix, wherein only identical matches and mismatches are considered, is typical. For proteins, this method usually involves two sets of parameters: a gap penalty and a substitution matrix assigning scores or probabilities to the alignment of each possible pair of amino acids based on the similarity of the amino acids' chemical properties and the evolutionary probability of the mutation. Most try to replicate evolution to get the most realistic alignment possible to best predict relations between sequences.Ī direct method for producing an MSA uses the dynamic programming technique to identify the globally optimal alignment solution. Each is usually based on a certain heuristic with an insight into the evolutionary process. There are various alignment methods used within multiple sequence to maximize scores and correctness of alignments. When choosing traces for a set of sequences it is necessary to choose a trace with a maximum weight to get the best alignment of the sequences. A trace is a set of realized, or corresponding and aligned, vertices that has a specific weight based on the edges that are selected between corresponding vertices. When determining the best suited alignments for each MSA, a trace is usually generated. Each of the graph edges has a weight based on a certain heuristic that helps to score each alignment or subset of the original graph.
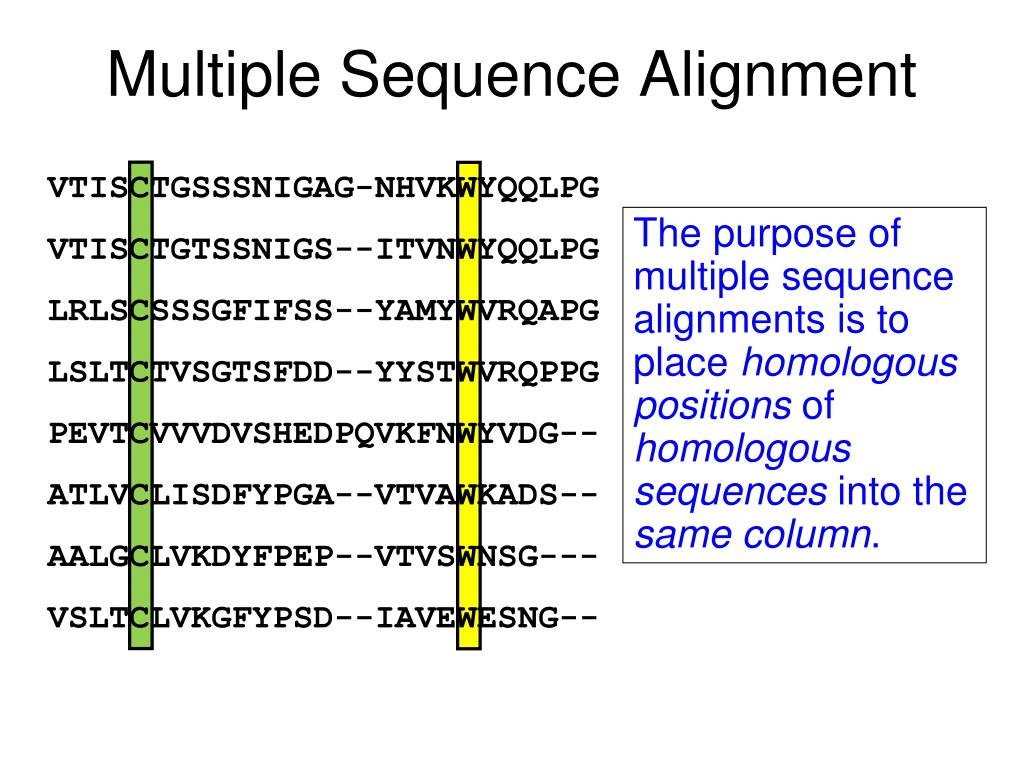
When finding alignments via graph, a complete alignment is created in a weighted graph that contains a set of vertices and a set of edges. Given m, remove all gaps.Ī general approach when calculating multiple sequence alignments is to use graphs to identify all of the different alignments. On the other hand, heuristic methods generally fail to give guarantees on the solution quality, with heuristic solutions shown to be often far below the optimal solution on benchmark instances. Most multiple sequence alignment programs use heuristic methods rather than global optimization because identifying the optimal alignment between more than a few sequences of moderate length is prohibitively computationally expensive. MSAs require more sophisticated methodologies than pairwise alignment because they are more computationally complex. Multiple sequence alignment is often used to assess sequence conservation of protein domains, tertiary and secondary structures, and even individual amino acids or nucleotides.Ĭomputational algorithms are used to produce and analyse the MSAs due to the difficulty and intractability of manually processing the sequences given their biologically-relevant length. Visual depictions of the alignment as in the image at right illustrate mutation events such as point mutations (single amino acid or nucleotide changes) that appear as differing characters in a single alignment column, and insertion or deletion mutations ( indels or gaps) that appear as hyphens in one or more of the sequences in the alignment. From the resulting MSA, sequence homology can be inferred and phylogenetic analysis can be conducted to assess the sequences' shared evolutionary origins. In many cases, the input set of query sequences are assumed to have an evolutionary relationship by which they share a linkage and are descended from a common ancestor. Multiple sequence alignment ( MSA) may refer to the process or the result of sequence alignment of three or more biological sequences, generally protein, DNA, or RNA. First 90 positions of a protein multiple sequence alignment of instances of the acidic ribosomal protein P0 (L10E) from several organisms.


 0 kommentar(er)
0 kommentar(er)
